A method for detergent-free isolation of membrane proteins in their local lipid environment
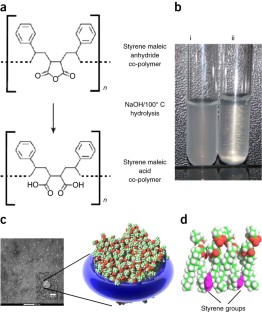
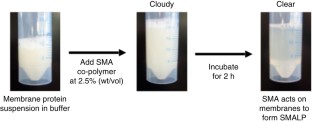
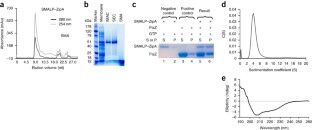
Despite the great importance of membrane proteins, structural and functional studies of these proteins present major challenges. A significant hurdle is the extraction of the functional protein from its natural lipid membrane. Traditionally achieved with detergents, purification procedures can be costly and time consuming. A critical flaw with detergent approaches is the removal of the protein from the native lipid environment required to maintain functionally stable protein. This protocol describes the preparation of styrene maleic acid (SMA) co-polymer to extract membrane proteins from prokaryotic and eukaryotic expression systems. Successful isolation of membrane proteins into SMA lipid particles (SMALPs) allows the proteins to remain with native lipid, surrounded by SMA. We detail procedures for obtaining 25 g of SMA (4 d); explain the preparation of protein-containing SMALPs using membranes isolated from Escherichia coli (2 d) and control protein-free SMALPS using E. coli polar lipid extract (1–2 h); investigate SMALP protein purity by SDS–PAGE analysis and estimate protein concentration (4 h); and detail biophysical methods such as circular dichroism (CD) spectroscopy and sedimentation velocity analytical ultracentrifugation (svAUC) to undertake initial structural studies to characterize SMALPs ( ∼ 2 d). Together, these methods provide a practical tool kit for those wanting to use SMALPs to study membrane proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
265,23 € per year
only 22,10 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Polyamine detergents tailored for native mass spectrometry studies of membrane proteins
Article Open access 14 September 2023
Modular detergents tailor the purification and structural analysis of membrane proteins including G-protein coupled receptors
Article Open access 28 January 2020
Lipid-polymer nanoparticles to probe the native-like environment of intramembrane rhomboid protease GlpG and its activity
Article Open access 30 August 2024
References
- Filmore, D. It's a GPCR world. Mod. Drug Discov.7, 24–28 (2004). CASGoogle Scholar
- Arinaminpathy, Y., Khurana, E., Engelman, D.M. & Gerstein, M.B. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov. Today14, 1130–1135 (2009). ArticleCASGoogle Scholar
- Seddon, A.M., Curnow, P. & Booth, P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta1666, 105–117 (2004). ArticleCASGoogle Scholar
- le Maire, M., Champeil, P. & Moller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta1508, 86–111 (2000). ArticleCASGoogle Scholar
- Gohon, Y. & Popot, J.-L. Membrane protein–surfactant complexes. Curr. Opin. Colloid Interface Sci.8, 15–22 (2003). ArticleCASGoogle Scholar
- Lin, S.-H. & Guidotti, G. Purification of membrane proteins. Methods Enzymol.463, 619–629 (2009). ArticleCASGoogle Scholar
- Damianoglou, A. et al. The synergistic action of melittin and phospholipase A2 with lipid membranes: development of linear dichroism for membrane-insertion kinetics. Protein Pept. Lett.17, 1351–1362 (2010). ArticleCASGoogle Scholar
- Charalambous, K., Miller, D., Curnow, P. & Booth, P.J. Lipid bilayer composition influences small multidrug transporters. BMC Biochem.9, 31 (2008). ArticleGoogle Scholar
- Wiener, M.C. & White, S.H. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of X-ray and neutron diffraction data. III. Complete structure. Biophys. J.61, 434–447 (1992). ArticleCASGoogle Scholar
- Simons, K. & Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol.3, a004697 (2011). ArticleGoogle Scholar
- Kellosalo, J., Kajander, T., Honkanen, R. & Goldman, A. Crystallization and preliminary X-ray analysis of membrane-bound pyrophosphatases. Mol. Membr. Biol.30, 64–74 (2013). ArticleGoogle Scholar
- Breyton, C., Tribet, C., Olive, J., Dubacq, J.P. & Popot, J.L. Dimer to monomer conversion of the cytochrome b6 f complex. Causes and consequences. J. Biol. Chem.272, 21892–21900 (1997). ArticleCASGoogle Scholar
- Esmann, M. Solubilized (Na+ + K+)-ATPase from shark rectal gland and ox kidney--an inactivation study. Biochim. Biophys. Acta857, 38–47 (1986). ArticleCASGoogle Scholar
- Popot, J.-L. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem.79, 737–775 (2010). ArticleCASGoogle Scholar
- Zoonens, M. & Popot, J.-L. Amphipols for each season. J. Membr. Biol.247, 759–796 (2014). ArticleCASGoogle Scholar
- Debnath, D.K., Basaiawmoit, R.V., Nielsen, K.L. & Otzen, D.E. The role of membrane properties in Mistic folding and dimerisation. Protein Eng. Des. Sel.24, 89–97 (2011). ArticleCASGoogle Scholar
- Denisov, I.G., Grinkova, Y.V., Lazarides, A.A. & Sligar, S.G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc.126, 3477–3487 (2004). ArticleCASGoogle Scholar
- Luthra, A., Gregory, M., Grinkova, Y.V., Denisov, I.G. & Sligar, S.G. Nanodiscs in the studies of membrane-bound cytochrome P450 enzymes. Methods Mol. Biol.987, 115–127 (2013). ArticleCASGoogle Scholar
- Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S.G. & Duong, F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J.26, 1995–2004 (2007). ArticleCASGoogle Scholar
- Leitz, A.J., Bayburt, T.H., Barnakov, A.N., Springer, B.A. & Sligar, S.G. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling nanodisc technology. Biotechniques40 601–2, 604, 606, passim (2006).
- Knowles, T.J. et al. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc.131, 7484–7485 (2009). ArticleCASGoogle Scholar
- Jamshad, M. et al. G-protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent. Biosci. Rep.35http://dx.doi.org/10.1042/BSR20140171 (2015).
- Tonge, S.R. & Tighe, B.J. Responsive hydrophobically associating polymers: a review of structure and properties. Adv. Drug Deliv. Rev.53, 109–122 (2001). ArticleCASGoogle Scholar
- Tonge, S., Stephen, T., Vincent, R. & Tighe, B.J. Dynamic surface activity of biological fluids and ophthalmic solutions. Cornea19, S133 (2000). ArticleGoogle Scholar
- Jamshad, M. et al. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res.8, 774–789 (2014). ArticleGoogle Scholar
- Scheidelaar, S. et al. Molecular model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers. Biophys. J.108, 279–290 (2015). ArticleCASGoogle Scholar
- Postis, V. et al. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta1848, 496–501 (2015). ArticleCASGoogle Scholar
- Gulati, S. et al. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J461, 269–278 (2014). ArticleCASGoogle Scholar
- Dörr, J.M. et al. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. USA111, 18607–18612 (2014). ArticleGoogle Scholar
- Paulin, S. et al. Surfactant-free purification of membrane protein complexes from bacteria: application to the staphylococcal penicillin-binding protein complex PBP2/PBP2a. Nanotechnology25, 285101 (2014). ArticleGoogle Scholar
- Orwick-Rydmark, M. et al. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett.12, 4687–4692 (2012). ArticleCASGoogle Scholar
- Long, A.R. et al. A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol.http://dx.doi.org/10.1186/1472-6750-13-41 (2013).
- Gomori, G., Colowick, S.P. & Kaplan, N.O. in Methods in Enzymology 138–148 (Academic Press, 1955). BookGoogle Scholar
- Lin, Y. Over-expression and Biophysical Characterisation of Membrane Proteins Solubilised in a Styrene Maleic Acid Polymer http://etheses.bham.ac.uk/1738/1/Lin_11_PhD.pdf (2011).
- Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J.78, 1606–1619 (2000). ArticleCASGoogle Scholar
- Fotiadis, D., Harder, D. & Fotiadis, D. Preparation of detergent-solubilized membranes from Escherichia coli. Protoc. Exchangehttp://dx.doi.org/10.1038/protex.2012.033 (2012).
- Panaretou, B. & Piper, P. in Yeast Protocols (ed. Evans, I.H.) 117–121 (Humana Press).
- Jamshad, M. et al. G-protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent. Biosci. Rep.35, 1–10 (2015). ArticleCASGoogle Scholar
- Scott, R.E. Plasma membrane vesiculation: a new technique for isolation of plasma membranes. Science194, 743–745 (1976). ArticleCASGoogle Scholar
- Del Piccolo, N., Placone, J., He, L., Agudelo, S.C. & Hristova, K. Production of plasma membrane vesicles with chloride salts and their utility as a cell membrane mimetic for biophysical characterization of membrane protein interactions. Anal. Chem.84, 8650–8655 (2012). ArticleCASGoogle Scholar
- Cohen, S., Ushiro, H., Stoscheck, C. & Chinkers, M. A native 170,000 epidermal growth factor receptor-kinase complex from shed plasma membrane vesicles. J. Biol. Chem.257, 1523–1531 (1982). CASPubMedGoogle Scholar
- Gulati, S. et al. Detergent free purification of ABC transporters. Biochem. J44, 1–24 (2014). Google Scholar
- Nordén, B., Rodger, A. & Dafforn, T. Linear Dichroism and Circular Dichroism: A Textbook on Polarized-light Spectroscopy (Royal Society of Chemistry, 2010).
- Burgess, S.A., Walker, M.L., Sakakibara, H., Oiwa, K. & Knight, P.J. The structure of dynein-c by negative stain electron microscopy. J. Struct. Biol.146, 205–216 (2004). ArticleCASGoogle Scholar
- Booth, D.S., Avila-Sakar, A. & Cheng, Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp.58, 3227 (2011). Google Scholar
- Grassucci, R.A., Taylor, D.J. & Frank, J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc.2, 3239–3246 (2007). ArticleCASGoogle Scholar
- Grassucci, R.A., Taylor, D. & Frank, J. Visualization of macromolecular complexes using cryo-electron microscopy with FEI Tecnai transmission electron microscopes. Nat. Protoc.3, 330–339 (2008). ArticleCASGoogle Scholar
- Scheres, S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–530 (2012). ArticleCASGoogle Scholar
- Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol.157, 38–46 (2007). ArticleCASGoogle Scholar
- Rico, A.I., Krupka, M. & Vicente, M. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J. Biol. Chem.288, 20830–20836 (2013). ArticleCASGoogle Scholar
- Hale, C.A., Rhee, A.C. & De Boer, P.A.J. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol.182, 5153–5166 (2000). ArticleCASGoogle Scholar
Acknowledgements
This work was supported by the Biotechnology and Biosciences Research Council (BBSRC; grants BB/J017310/1, BB/H016309/1, BB/I005579/1, BB/I020349/1, BB/K004441/1 and BB/M018261/1 to T.R.D.) and the Wellcome Trust (ref. 091322/7/10/Z to V.L.G.P.). S.P.M. is supported by an MRC Career Development Award (G100567). We dedicate this work to the late Professor Stephen A. Baldwin.
Author information
- Tim J Knowles & Pooja Sridhar Present address: Present address: Department of Biosciences, University of Birmingham, Birmingham, UK.,
- Sarah C Lee, Tim J Knowles and Vincent L G Postis: These authors contributed equally to this work.
Authors and Affiliations
- School of Biosciences, University of Birmingham, Birmingham, UK Sarah C Lee, Mohammed Jamshad, Rosemary A Parslow, Yu-pin Lin, Michael Overduin & Timothy R Dafforn
- School of Cancer Sciences, University of Birmingham, Birmingham, UK Tim J Knowles & Pooja Sridhar
- School of Biomedical Sciences, Faculty of Biological Sciences, University of Leeds, Leeds, UK Vincent L G Postis, Adrian Goldman & Stephen P Muench
- Biomedicine Research Group, Faculty of Health and Social Sciences, Leeds Beckett University, Leeds, UK Vincent L G Postis
- Department of Biosciences, Division of Biochemistry, University of Helsinki, Helsinki, Finland Adrian Goldman
- Department of Biochemistry, Faculty of Medicine & Dentistry, University of Alberta, Edmonton, Alberta, Canada Michael Overduin
- Sarah C Lee








